The Basics of Encoding Categorical Data for Predictive Models
Thomas Yokota asked a very straight-forward question about encodings for categorical predictors: “Is it bad to feed it non-numerical data such as factors?” As usual, I will try to make my answer as complex as possible.
(I’ve heard the old wives tale that eskimos have 180 different words in their language for snow. I’m starting to think that statisticians have at least as many ways of saying “it depends”)
BTW, we cover this in Sections 3.6, 14.1 and 14.7 of the book.
My answer: it depends on the model. Some models can work with categorical predictors in their nature, non-numeric encodings. Trees, for example, can usually partition the predictor into distinct groups. For a predictor X with levels a through e, a split might look like
if X in {a, c, d} the class = 1
else class = 2Rule-based models operate similarly.
Naive Bayes models are another method that does not need to re-encode the data. In the above example, the frequency distribution of the predictor is computed overall as well as within each of the classes (a good example of this is in Table 13.1 for those of you that are following along).
However, these are the exceptions; most models require the predictors to be in some sort of numeric encoding to be used. For example, linear regression required numbers so that it can assign slopes to each of the predictors.
The most common encoding is to make simple dummy variables. The there are C distinct values of the predictor (or levels of the factor in R terminology), a set of C - 1 numeric predictors are created that identify which value that each data point had.
These are called dummy variables. Why C - 1 and not C? First, if you know the values of the first C - 1 dummy variables, you know the last one too. It is more economical to use C - 1. Secondly, if the model has slopes and intercepts (e.g. linear regression), the sum of all of the dummy variables wil add up to the intercept (usually encoded as a “1”) and that is bad for the math involved.
In R, a simple demonstration for the example above is:
> pred1 <- factor(letters[1:5])
> pred1
[1] a b c d e
Levels: a b c d eThe R function model.matrix is a good way to show the encodings:
> model.matrix(~pred1)
(Intercept) pred1b pred1c pred1d pred1e
1 1 0 0 0 0
2 1 1 0 0 0
3 1 0 1 0 0
4 1 0 0 1 0
5 1 0 0 0 1
attr(,"assign")
[1] 0 1 1 1 1
attr(,"contrasts")
attr(,"contrasts")$pred1
[1] "contr.treatment"A column for the factor level a is removed (since it excludes the first level of the factor). This approach goes by the name of “full-rank” encoding since the dummy variables do not always add up to 1.
We discuss different encodings for predictors in a few places but fairly extensively in Section 12.1. In that example, we have a predictor that is a date. Do we encode that as the day or the year (1 to 365) and include it as a numeric predictor? We could also add in predictors for the day of the week, the month, the season etc. There are a lot of options. This question of feature engineering is important. You want to find the encoding that captures the important patterns in the data. If there is a seasonal effect, the encoding should capture that information. Exploratory visualizations (perhaps with lattice or ggplot2) can go a long way to figuring out good ways to represent these data.
Some of these options result in ordered encodings, such as the day of the week. It is possible that the trends in the data are best exposed if the ordering is preserved. R does have a way for dealing with this:
> pred2 <- ordered(letters[1:5])
> pred2
[1] a b c d e
Levels: a < b < c < d < eSimple enough, right? Maybe not. If we need a numeric encoding here, what do we do?
There are a few options. One simple way is to assign “scores” to each of the levels. We might assign a value of 1 to a and think that b should be twice that and c should be four times that and so on. It is arbitrary but there are whole branches of statistics dedicated to modeling data with (made up) scores. Trend tests are one example.
If the data are ordered, one technique is to create a set of new variables similar to dummy variables. However, their values are not 0/1 but are created to reflect the possible trends that can be estimated.
For example, if the predictor has two ordered levels, we can’t fit anything more sophisticated to a straight line. However, if there are three ordered levels, we could fit a linear effect as well as a quadratic effect and so on. There are some smart ways to do this (google “orthogonal polynomials” if you are bored).
For each ordered factor in a model, R will create a set of polynomial scores for each (we could use the fancy label of “a set of basis functions” here). For example:
> model.matrix(~pred2)
(Intercept) pred2.L pred2.Q pred2.C pred2^4
1 1 -0.6325 0.5345 -3.162e-01 0.1195
2 1 -0.3162 -0.2673 6.325e-01 -0.4781
3 1 0.0000 -0.5345 -4.096e-16 0.7171
4 1 0.3162 -0.2673 -6.325e-01 -0.4781
5 1 0.6325 0.5345 3.162e-01 0.1195
attr(,"assign")
[1] 0 1 1 1 1
attr(,"contrasts")
attr(,"contrasts")$pred2
[1] "contr.poly"Here, “L” is for linear, “Q” is quadratic and “C” is cubic and so on. There are five levels of this factor and we can create four new encodings. Here is a plot of what those encodings look like:
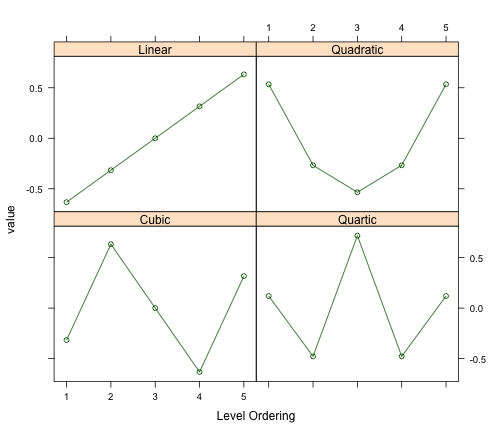
The nice thing here is that, if the underlying relationship between the ordered factor and the outcome is cubic, we have a feature in the data that can capture that trend.
One other way of encoding ordered factors is to treat them as unordered. Again, depending on the model and the data, this could work just as well.
(This article was originally posted at http://appliedpredictivemodeling.com)